Mac; New Terminal: Alt+Shift+R: Shift+Option+R: Move Focus to Terminal: Alt+Shift+M. Ctrl+Option+O: Tools Menu: Alt+Shift+T: Ctrl+Option+T: Help Menu: Alt+Shift+H.
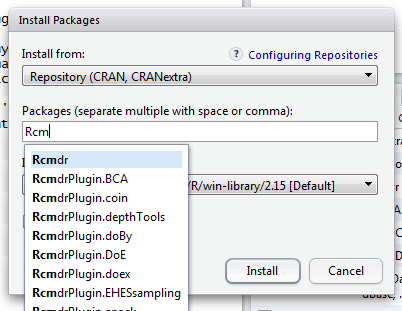
- Choose the Recovery Drive to launch Mac. Internet Recovery. If there is an opportunity to connect your Mac online, you really can attempt Internet Recovery. Just as its name shows, you need to get your Mac online. Here is how to use Internet Recovery. Reboot Mac and tap 'Option' 'Command+R' when you hear the startup music.
- Press Options - Command - R to upgrade to the latest version of macOS that's compatible with your Mac. Press Shift - Option - Command - R to install the version of macOS that came with your Mac, or the version closest to it that's still available. How to check your startup disk with Disk Utility in macOS Recovery mode.

This guide was developed as way for me to take notes as I tried to convert a number of routines into a more easilly managed 'package'. It is geared to the Mac user who is clueless with respect to Windows and can stumble around (with some limited success) in the Unix part of the Mac.
Tool Option In R For Macs
An equally frustrated pair of users created this guide for Linux, Unix and Windows users. (Unfortunately, their page has a link for how to build packages for Macs and it refers back to this page. So, I can just recommend reading that page for other hints.)
Tool Option In R For Mac Os
Also, consult the R for Mac OS FAQ page which is the definitive resource for building packages, but does assume more ability than I seem to have to follow it completely.
Notes:
- In the various tutorials, one will see the
R CMD X
line. This means that one should execute this line while using the console (using X-11), not from the R Gui. Although perhaps obvious to everyone else, this was not obvious to me. - Files will be created from various routines (e.g., prompt), that will need to be moved to where you are building your packages.
- The home location for R packages from the Mac finder on my machine is 'desktop/Macintosh HD/Library/Frameworks/R.framework/Resources/library/'. Within library you will see all the packages that you have downloaded, and installed. Examining some of those, e.g., the sem package, helps one understand what goes where.
- the 'tools' package should be installed.
Making the package -- the first time
- package.skeleton(name = 'anRpackage', list, environment = .GlobalEnv, path = '.', force = FALSE) will createthe appropriate files for a package.
So, for instance in making up the psych package, the first step is to make the skeleton. This list is strung out to make it more readable. Note that I include a dummy function (psych) with the same name as the package. This allows one to get help by merely asking ?psych. (This seems to have changed in version 2.3.1 and now the skeleton automatically makes up a file with the name of the package - This then creates a new folder (at the root directory level) that has draft DESCRIPTION, INDEX, AND CONTENT files, and three folders: R, man, and scr, in addition to a 'read and delete me file that says '
- The R folder/directory contains each function listed in the call to the skeleton function created as an individual file with the suffix R. e.g., alpha.scale.R, omega.R, etc. Corrections to your code can be done directly to these files.
- The man folder/directory has has a number of short files that include basic descriptions of each function. These need to be edited to provide basic information for the help files. Particular, the ~kwd1 and ~kwd2 should match the Contents file and use keywords from the KEYWORDS.db.
Note, that the files in the man folder are written in a .Rd format. This format is used by R to create HTML and LaTex files. Although it is tempting to edit the html files that will eventually appear in the library, it turns out that one needs to learn to edit the Rd formated files. See section 2 of the package manual. The process of building the package takes these Rd files and converts them to HTML as well as Latex and (perhaps) pdf. As Rossi points out, it is vital to write clear documentation for your functions. This is, of course, the least fun part of program development, but essential.
The syntax of these .Rd files is moderately straight forward. The % mark signifies a comment. All commands start with a and are defined within { }.
- We now have a folder that can be built. But first, we need to correct the description file. (Without fixing the description that comes out of package.skeleton, build fails.) In addition, as far as I can tell, periods are not allowed in the Description.
An example description file
- At this point you can go to the X11 window and use the R CMD build psych command.
- Building will produce a tar.gz file that can be loaded using the package installer. The package manager, in turn, will take that file and create the appropriate HTML and pdf help files.
The package installer can load local packages (as source files) from your root directory. The installer then makes the html and pdf help files based upon your .Rd files.
If corrections to the package are to be made, they should be done in the original files in the original folder and then the build sequence can be followed again.
- R CMD check psych will check all the syntax of your files, including the help files. Its diagnostics will appear on the screen as well as in the psych.R.check folder. Fixing all the complaints will take a while, but is necessary to make a package that can be sent to CRAN.
Adding corrections and additions to the packageIf you want to add any new functions to the package, you can save the function as foo.R in the R folder of the package and then add the appropriate help file (.Rd) in the man folder/directory. To create the new help file, use the prompt(foo) command. This will write foo.Rd into the man directory (folder).
After adding a new function (foo.R) to the foo.package directory and running prompt(foo.R) to create the foo.Rd file to be put in the foo.man directory, run R CMD check foo.package until no errors occur. (Be patient.)
Examine the Contents file and give keywords that match those found in the KEYWORDS.db file for R. Also add 1 line descriptions to each function. The index is created by the package manager, and uses the one line desciption files.
Sharing your package -- repositories and CRANWe now have a workable package. This can be tested and uploaded to Cran, or can be installed in a local 'repository' for further testing and development. Note, that if the package is just written in R, it is loaded as source rather than as binary.
The use of repositories has been discussed in Rnews by Brian Ripley. Repositories are meant for development of department or university wide packages that might not be of general interest. I have been doing this for the psych package, trying to maintain both a PC and a Mac version. The Mac version has the requirement (according to the Mac FAQ): 'A Mac OS X specific requirements, is that a prebuilt package is assumed to be named (and accordingly archived and compressed) as package_name.tgz. On Windows, for example, packages come in a zipped format.'
To do this, we need to create a number of folders (directories if using a PC) and files. The basic default location for my repository is
- http://personality-project.org/r/
This has a folder (directory) of src which contains - contrib which contains at least 3 files:
- PACKAGES
- This file is a listing of the packages in the repository.
- for me, with one file it is merely
Package: psych
Version: 1.0-7
Depends: NA
- PACKAGES.html
A basic html file with a table describing the various packages. - psych_1.0-7.tar.gz the actual package
- as well as the folders:
- Descriptions which contains
- psych.html
- Descriptions which contains
- PACKAGES
Appropriate read permissions need to be added to these files and folders to allow others to access them.
Once this is done once, it is then possible to do direct edits to this set of files to make improvements to the package. However, this is a bad idea. It is much better to make the corrections to the package master folder (the pre built one, not the installed one), for that corrects the help files as well.
- Files to fix
- actual program corrections may be done to the file 'psych' in the subdirectory (folder) R. When the package is rebuilt, these corrections will be added to the package.
- Corrections to the help documentation can be done to the files in the folder Help. However, these are not actually part of the package, unless the corrections are done to the .Rd files.
- HTML help files are found in the HTML folder. These are the files that are used when asking for help or from the index. I had thought these are the most important to revise, and although this can be done, it is better to spend the time fixing the .Rd files and the remaking the package.
Once the habit of correcting .Rd files and function.R files becomes routinized, then the rebuilding operation is fairly straight forward.
R CMD check psych is a very useful (although tedious) operation. This will work through mistake after mistake in the .Rd files. Correct these!
Using X-11 to finish
- After doing corrections to the various files, one can make an updated tgz package by using X-11
- tar --create -f psych.1.0-7.tar psych (takes the folder psych and compresses it)
- gzip psych.1.0-7.tar (takes the file and gzips it)
- zip -r psych psych (makes a zip file for pc users)
- then, it is necessary to set the correct permissions for these files so that they can be read.
Then, finally, we move the psych.1.0-7.tgz file to the PACKAGEs folder and it is ready to be installed by other Mac users.
I think it is preferred to actually do this to the result of the build operation rather than the folder inside the library.
To allow PC users to use the package, they need to be able to download the zip file. Thus, they need to be able to find it! I have created a folder r/bin/pcos that has the psych package. Presumably some introductory remarks similar to those found in the Mac package structure would be helpful.
How to set up a repository using BioConductor is discussed by the bioconductor folk, (but I am unable to make this work).http://www.maths.lth.se/help/R/.R/library/reposTools/doc/reposServer.pdf
With my package 'psych' installed on the personality-project.org server, one can install it by using the R-package installer and specifying 'other respository' with the address http://personality-project.org/r . It magically works its way down the file structure to get the appropriate package.
part of a short guide to R
Version of June 13, 2006
William Revelle
Department of Psychology
Northwestern University - http://personality-project.org/r/
